Gamma GLM
Psychological Sciences
Filippo Gambarota
University of Padova
Last modified: 08-12-2025
Gamma Distribution
Gamma distribution
The Gamma distribution has several :parametrizations. One of the most common is the shape-scale parametrization:
\[ f(x;k,\theta )={\frac {x^{k-1}e^{-x/\theta }}{\theta ^{k}\Gamma (k)}} \]
Where \(\theta\) is the scale parameter and \(k\) is the shape parameter.
Gamma distribution
Gamma \(\mu\) and \(\sigma^2\)
The mean and variance are defined as:
- \(\mu = k \theta\) and \(\sigma^2 = k \theta^2\) with the shape-scale parametrization
- \(\mu = \frac{\alpha}{\beta}\) and \(\frac{\alpha}{\beta^2}\) with the shape-rate parametrization
Another important quantity is the coefficient of variation defined as \(\frac{\sigma}{\mu}\) or \(\frac{1}{\sqrt{k}}\) (or \(\frac{1}{\sqrt{\alpha}}\)).
Gamma distribution
Again, we can see the mean-variance relationship:
Gamma parametrization
To convert between different parametrizations, you can use the gamma_params() function:
gamma_params <- function(
mean = NULL,
sd = NULL,
shape = NULL,
scale = NULL,
rate = NULL,
skew = NULL,
cv = NULL
) {
pars <- as.list(environment())
spars <- pars[!sapply(pars, is.null)]
if (length(spars) != 2) {
stop("Please provide exactly two parameters from a supported pair.")
}
is_pair <- function(x, pair) identical(sort(x), sort(pair))
compute <- function(shape, scale) {
rate <- 1 / scale
mean <- shape * scale
sd <- sqrt(shape * scale^2)
cv <- 1 / sqrt(shape)
skew <- 2 / sqrt(shape)
list(
shape = shape,
rate = rate,
scale = scale,
cv = cv,
skew = skew,
mean = mean,
sd = sd
)
}
if (is_pair(names(spars), c("mean", "sd"))) {
mean <- spars$mean
sd <- spars$sd
cv <- sd / mean
shape <- 1 / cv^2
scale <- mean / shape
} else if (is_pair(names(spars), c("mean", "shape"))) {
mean <- spars$mean
shape <- spars$shape
scale <- mean / shape
} else if (is_pair(names(spars), c("mean", "cv"))) {
mean <- spars$mean
cv <- spars$cv
shape <- 1 / cv^2
scale <- mean * cv^2
} else if (is_pair(names(spars), c("shape", "rate"))) {
shape <- spars$shape
scale <- 1 / spars$rate
} else if (is_pair(names(spars), c("shape", "scale"))) {
shape <- spars$shape
scale <- spars$scale
} else if (is_pair(names(spars), c("mean", "skew"))) {
mean <- spars$mean
skew <- spars$skew
shape <- 4 / skew^2
scale <- mean / shape
} else {
stop("combination not implemented!")
}
compute(shape, scale)
}Gamma parametrization
Gamma parametrization
Understanding parameters
\(\mu\) and \(\sigma\) parametrization
- Using the
gamma_params()function we can think in terms of \(\mu\) and \(\sigma\) and generate the right parameters (e.g., shape and rate). - Let’s simulate observations from a Gamma distribution with \(\mu = 500\) and \(\sigma = 200\)
\(\mu\) and \(\sigma\) parametrization
Then we can fit an intercept-only model with the Gamma family and a log link function. You have to specify the link because the default is inverse.
fam <- Gamma(link = "log")
dat <- data.frame(y)
fit0 <- glm(y ~ 1, family = fam, data = dat)
summary(fit0)#>
#> Call:
#> glm(formula = y ~ 1, family = fam, data = dat)
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) 6.215218 0.004027 1543 <2e-16 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> (Dispersion parameter for Gamma family taken to be 0.1621637)
#>
#> Null deviance: 1646.3 on 9999 degrees of freedom
#> Residual deviance: 1646.3 on 9999 degrees of freedom
#> AIC: 133274
#>
#> Number of Fisher Scoring iterations: 4\(\mu\) and \(\sigma\) parametrization
\(\beta_0\) is the \(\mu\) of the Gamma distribution. We need to apply the inverse (exp) to get the original scale:
#> mean mean_true b0.(Intercept)
#> 500.305 500.000 500.305as always you can use the fam() object if you are not sure about the link functions as fam$linkinv(coef(fit0)[1])
\(\mu\) and \(\sigma\) parametrization
Now let’s simulate the difference between two groups. Again fixing the \(\mu_0 = 500\), \(\mu_1 = 600\) and a common \(\sigma = 200\). Let’s plot the empirical densities:
\(\mu\) and \(\sigma\) parametrization
Using the group variable as dummy-coded, \(\beta_0 = \mu_0\) and \(\beta_1 = \mu_1 - \mu_0\). Note that we are in the log scale.
ns <- 1e4
m0 <- 500
m1 <- 600
s <- 200
# parameters, log link
b0 <- log(m0)
b1 <- log(m1) - log(m0) # equivalent to log(m1 / m0)
x <- rep(c(0, 1), each = ns/2)
lp <- b0 + b1 * x # linear predictor
mu <- exp(lp) # inverse exp link
gm <- gamma_params(mean = mu, sd = s)
y <- rgamma(ns, shape = gm$shape, scale = gm$scale)
dat <- data.frame(y, x)\(\mu\) and \(\sigma\) parametrization
Let’s see the simulated data:
\(\mu\) and \(\sigma\) parametrization
Now we can fit the model and extract the parameters:
#>
#> Call:
#> glm(formula = y ~ x, family = fam, data = dat)
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) 6.214232 0.005265 1180.39 <2e-16 ***
#> x 0.175887 0.007445 23.62 <2e-16 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> (Dispersion parameter for Gamma family taken to be 0.1385776)
#>
#> Null deviance: 1506.1 on 9999 degrees of freedom
#> Residual deviance: 1428.9 on 9998 degrees of freedom
#> AIC: 133781
#>
#> Number of Fisher Scoring iterations: 4\(\mu\) and \(\sigma\) parametrization
\(\beta_0\) is the mean of the first group and \(\beta_1\) is the \(\log(\mu_1/\mu_0)\) or the difference \(\log(\mu_1) - log(\mu_0)\)
\(\mu\) and \(\sigma\) parametrization
The other estimated parameter is the dispersion that is defined as the inverse of the shape. We have not a single shape but the average is roughly similar to the true value.
\(\mu\) and shape parametrization
This is common in brms and other packages1. The \(\mu\) is the same as before and the shape (\(\alpha\)) determine the skewness of the distribution. For the Gamma, the skewness is calculated as \(\frac{2}{\sqrt{\alpha}}\).
To generate data, we calculate the scale (\(\theta\)) as \(\frac{\mu}{\alpha}\) (remember that \(\mu = \alpha\theta\))
\(\mu\) and shape parametrization
the expected skewness is 2/sqrt(shape) 0.6324555 and is similar to the value computed on the simulated data
as \(\alpha\) increase, the Gamma distribution is less skewed and approaches a Gaussian distribution. When \(\mu = \alpha\) the distribution already start to be pretty Gaussian
Skewness - \(\alpha\) relationship
We can plot the function that determine the skewness of the Gamma fixing \(\mu\) and varying \(\alpha\):
Skewness - \(\alpha\) relationship
Compared to the \(\mu\)-\(\sigma\) method, here we fix the skewness and \(\mu\), thus the \(\hat \sigma\) will differ when \(\mu\) change but the skewness is the same. The opposite is also true.
mu <- c(50, 80)
# mu-shape parametrization
y1 <- rgamma(1e6, shape = 10, scale = mu[1]/10)
y2 <- rgamma(1e6, shape = 10, scale = mu[2]/10)
# mu-sigma parametrization
gm <- gamma_params(mean = mu, sd = c(20, 20))
x1 <- rgamma(1e6, shape = gm$shape[1], scale = gm$scale[1])
x2 <- rgamma(1e6, shape = gm$shape[2], scale = gm$scale[2])
par(mfrow = c(1,2))
plot(density(y1), lwd = 2, main = latex("\\mu and \\alpha parametrization"), xlab = "x", xlim = c(0, 250))
lines(density(y2), col = "firebrick", lwd = 2)
legend("topright",
legend = c(latex("\\mu = %s, \\alpha = %s, \\hat{\\sigma} = %.0f, sk = %.2f", mu[1], 10, sd(y1), psych::skew(y1)),
latex("\\mu = %s, \\alpha = %s, \\hat{\\sigma} = %.0f, sk = %.2f", mu[2], 10, sd(y2), psych::skew(y1))),
fill = c("black", "firebrick"))
hatshape <- c(gamma_shape(x1, "invskew"), gamma_shape(x2, "invskew"))
plot(density(x1), lwd = 2, main = latex("\\mu and \\sigma parametrization"), xlab = "x", xlim = c(0, 250))
lines(density(x2), col = "firebrick", lwd = 2)
legend("topright",
legend = c(latex("\\mu = %s, \\sigma = %s, \\hat{\\alpha} = %.0f, sk = %.2f", mu[1], 20, hatshape[1], psych::skew(x1)),
latex("\\mu = %s, \\sigma = %s, \\hat{\\alpha} = %.0f, sk = %.2f", mu[2], 20, hatshape[2], psych::skew(x2))),
fill = c("black", "firebrick"))Coefficient of variation
The coefficient of variation \(\frac{\sigma}{\mu} = \frac{1}{\sqrt{\alpha}}\) is constant under the \(\mu\)-\(\alpha\) parametrization while can be different under the \(\mu\)-\(\sigma\) one when \(\alpha\) or \(\sigma\) is fixed across conditions.
# mu-shape
c(cv(y1), cv(y2))#> [1] 0.3165037 0.3158017# mu-sigma
c(cv(x1), cv(x2))#> [1] 0.3997695 0.2499677The \(\alpha\) parameter allow to control the coefficient of variation.
\(\mu\) and \(\sigma\) relationship
See https://civil.colorado.edu/~balajir/CVEN6833/lectures/GammaGLM-01.pdf. The \(\sigma = \frac{\mu}{\sqrt{\alpha}}\).
\(\mu\) and \(\sigma\) relationship
As noted by Agresti (2015), fixing \(\alpha\) and varying \(\mu\), the coefficient of variation will be constant and the standard deviation \(\sigma\) increase proportionally with \(\mu\). Given that \(\sigma = \frac{\mu}{\sqrt{\alpha}}\):
mu1 <- 20
mu2 <- 40
shape <- 10 # alpha
y1 <- rgamma(1e5, shape = shape, scale = mu1/shape)
y2 <- rgamma(1e5, shape = shape, scale = mu2/shape)
c(mean(y1), mean(y2))
#> [1] 20.02286 40.03831
c(sd(y1), sd(y2)) # sd increase
#> [1] 6.313313 12.699355
c(cv(y1), cv(y2)) # cv is constant
#> [1] 0.3153053 0.3171801
c(psych::skew(y1), psych::skew(y2)) # skewness is similar
#> [1] 0.6452390 0.6335816Example: the Simon effect
The Simon effect is the difference in accuracy or reaction time between trials in which stimulus and response are on the same side and trials in which they are on opposite sides, with responses being generally slower and less accurate when the stimulus and response are on opposite sides.
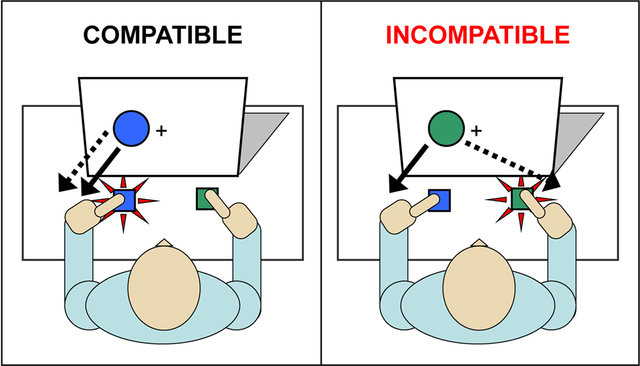
Source: Wildenberg et al. (2010)
Example: the Simon effect
Let’s import the data/simon.rda file1. You can use the load() function:
Example: the Simon effect
For simplicity, let’s consider only a single subject (submission_id: 7432), otherwise the model require including random effects. We also exclude strange trials with RT > 2500 ms.
simon <- filter(simon,
submission_id == 7432,
RT < 2500)Example: the Simon effect
Let’s plot the reaction times. Clearly the two distributions are right-skewed with a difference in location (\(\mu\)). The shape also differs between thus also the skewness is probably different:
ggplot(simon, aes(x = RT, fill = condition)) +
geom_density(alpha = 0.7)Example: the Simon effect
Let’s see some summary statistics. We see the difference between the two conditions.
#> $congruent
#> mean sd skew cv
#> 504.1818182 81.7038670 0.5328521 0.1620524
#>
#> $incongruent
#> mean sd skew cv
#> 564.0312500 123.4188852 2.8702013 0.2188157Example: the Simon effect
Given that we modelling the difference in \(\mu\), this is the expected difference. We are working on the log scale, thus the model is estimating the log difference or the log ratio.
Example: the Simon effect
Let’s fit the model:
#>
#> Call:
#> glm(formula = RT ~ condition, family = Gamma(link = "log"), data = simon)
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) 6.22294 0.02625 237.053 < 2e-16 ***
#> conditionincongruent 0.11217 0.03580 3.134 0.00218 **
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> (Dispersion parameter for Gamma family taken to be 0.03790215)
#>
#> Null deviance: 4.1148 on 118 degrees of freedom
#> Residual deviance: 3.7439 on 117 degrees of freedom
#> AIC: 1424.4
#>
#> Number of Fisher Scoring iterations: 4Example: the Simon effect
Plotting the results:
plot(ggeffect(fit))Example: the Simon effect
The main parameter of interest here is the \(\beta_1\) representing the difference in \(\mu\). We can interpret \(\exp(\beta_1) = 1.119\) as the multiplicative increase in RT when moving from congruent to incongruent condition. In the RT scale, we have a difference of 59.8494318. Remember that the statistical test is performed on the link-function scale.
emmeans(fit, pairwise ~ condition)$contrast
#> contrast estimate SE df t.ratio p.value
#> congruent - incongruent -0.112 0.0358 117 -3.134 0.0022
#>
#> Results are given on the log (not the response) scale.
emmeans(fit, pairwise ~ condition, type = "response")$contrast
#> contrast ratio SE df null t.ratio p.value
#> congruent / incongruent 0.894 0.032 117 1 -3.134 0.0022
#>
#> Tests are performed on the log scaleMore about the Gamma distribution
I have written more notes about the intepretation of Gamma parameters especially for simulations: https://filippogambarota.github.io/notes/understanding-gamma/