probrep = function(
nn, # sample size of both the initial and replication experiment
gamma, # prior probability of the hypothesis
theta0, # success probability under the null hypothesis
theta1, # success probability under the null hypothesis
alpha # significance cutoff
){
x1range = seq(0,nn) # possible values of original experiment
pvalue1 = pH0 = rep(NA,nn+1) # initialization
# type two error estimation via quick simulation
MM = 10000
xx.under.alt = rbinom(MM,nn,theta1)
pvalue.alt = NA
for (mm in 1:MM){
pvalue.alt[mm] = binom.test(xx.under.alt[mm],nn,theta0,alternative="two.sided")$p.value
}
beta = mean( pvalue.alt > .05) # type one error in both studies
#
for (xx in x1range){
# mapping between number of successes and pvalue in original ezxperiment
pvalue1[xx] =
binom.test(xx,nn,theta0,alternative="two.sided")$p.value
# probability of null hypothesis given first experiment
pH0[xx] = gamma * dbinom(xx,nn,theta0) /
( gamma * dbinom(xx,nn,theta0) + (1-gamma) * dbinom(xx,nn,theta1))
}
# set tiny p-values to e-20 for visualization purposes
pvalue1[is.na(pvalue1)] = 0
pH0[is.na(pH0)] = 0
# separate rejections
xx.reject = x1range[ pvalue1 < .05 ] + 1
xx.retain = x1range[ pvalue1>=.05 ] + 1
pvalue.reject = pvalue1[xx.reject]
pvalue.retain = pvalue1[xx.retain]
pH0.reject = pH0[xx.reject]
pH0.retain = pH0[xx.retain]
# browser()
# compute replication probabilitie
prep.reject = (alpha/2) * pH0[xx.reject] + (1-beta) * (1-pH0[xx.reject])
prep.retain = (1-alpha) * pH0[xx.retain] + beta * (1-pH0[xx.retain])
return(list(
reject = na.omit(data.frame(prep.reject,pvalue.reject,pH0.reject)),
retain = na.omit(data.frame(prep.retain,pvalue.retain,pH0.retain))))
}Probability of Replication
Probability of replication of a Binomial test for simple versus simple hypothesis. Inspired by Miller’s paper.
Model: \(x\) is Binomial \(\theta\). There are two possible values of \(\theta\) the null value \(\theta_0\) and the alternative \(\theta_1\). Both the original and replicated experiment have size \(n\). We know \(\gamma\).
The logic is this:
-- compute the power \((1-\beta\)) of the studies (it is the same for both) by simulation
-- for ever possible \(x\) in \(0, \ldots, n\) compute the p-value and the posterior probability of the null \(\theta=\theta_0\)
-- if the \(x\) leads to a rejection compute the probability of replication as \(\alpha/2 * p(\theta_0|x) + (1-\beta) (1-p(\theta_0|x))\)
-- if the \(x\) does not lead to a rejection compute the probability of replication as \((1-\alpha) * p(\theta_0|x) + \beta (1-p(\theta_0|x))\)
Function to compute the probability of replication.
probrep.out = probrep(nn=20,gamma=.5,theta0=.1,theta1=.2,alpha=0.05)
plot(probrep.out$reject$pvalue.reject,
probrep.out$reject$prep.reject,
ylab="probability of replication",
xlab="p-value of original study")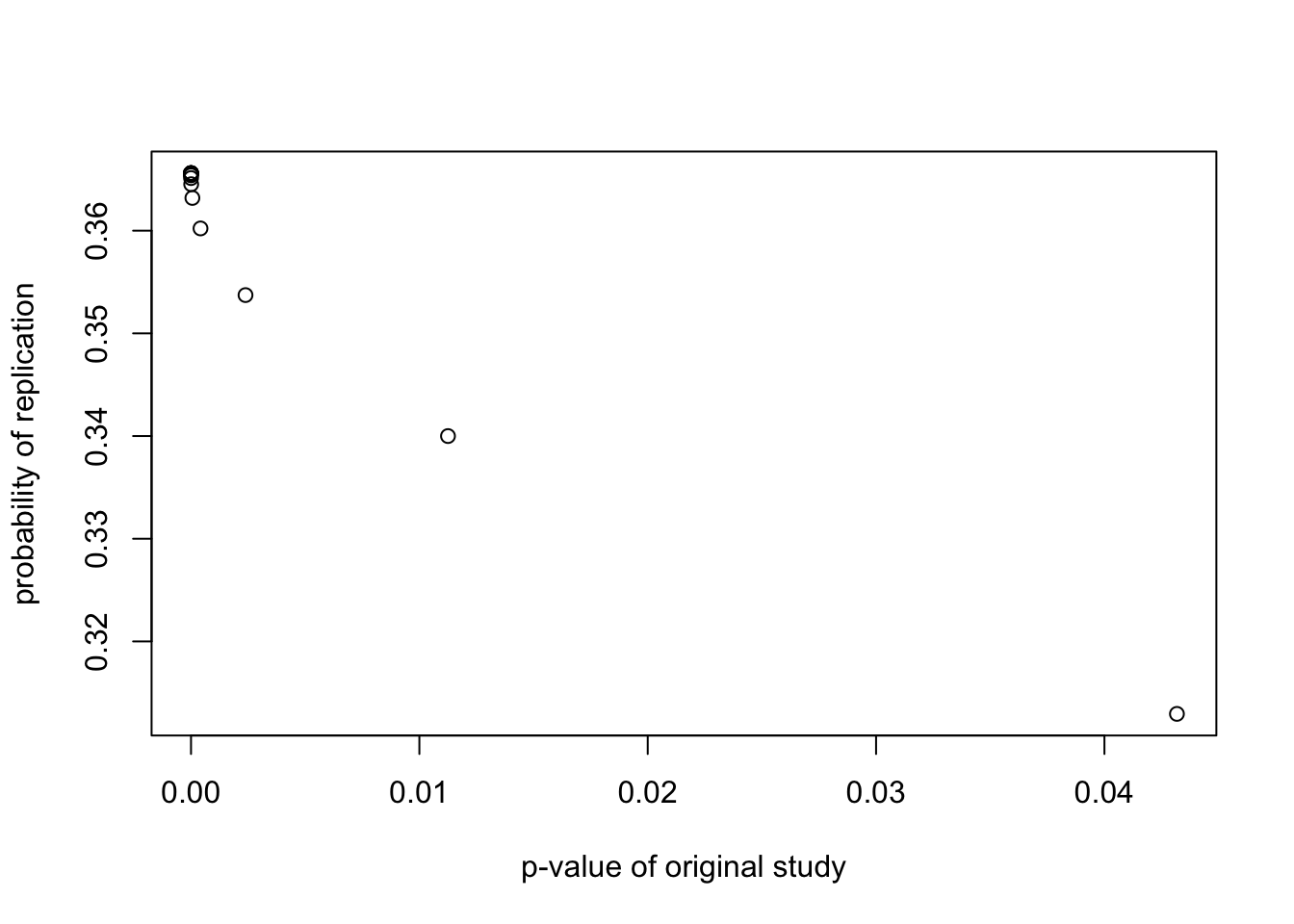
plot(probrep.out$reject$pvalue.reject,
probrep.out$reject$pH0.reject,
ylab="probability of null hypothesis",
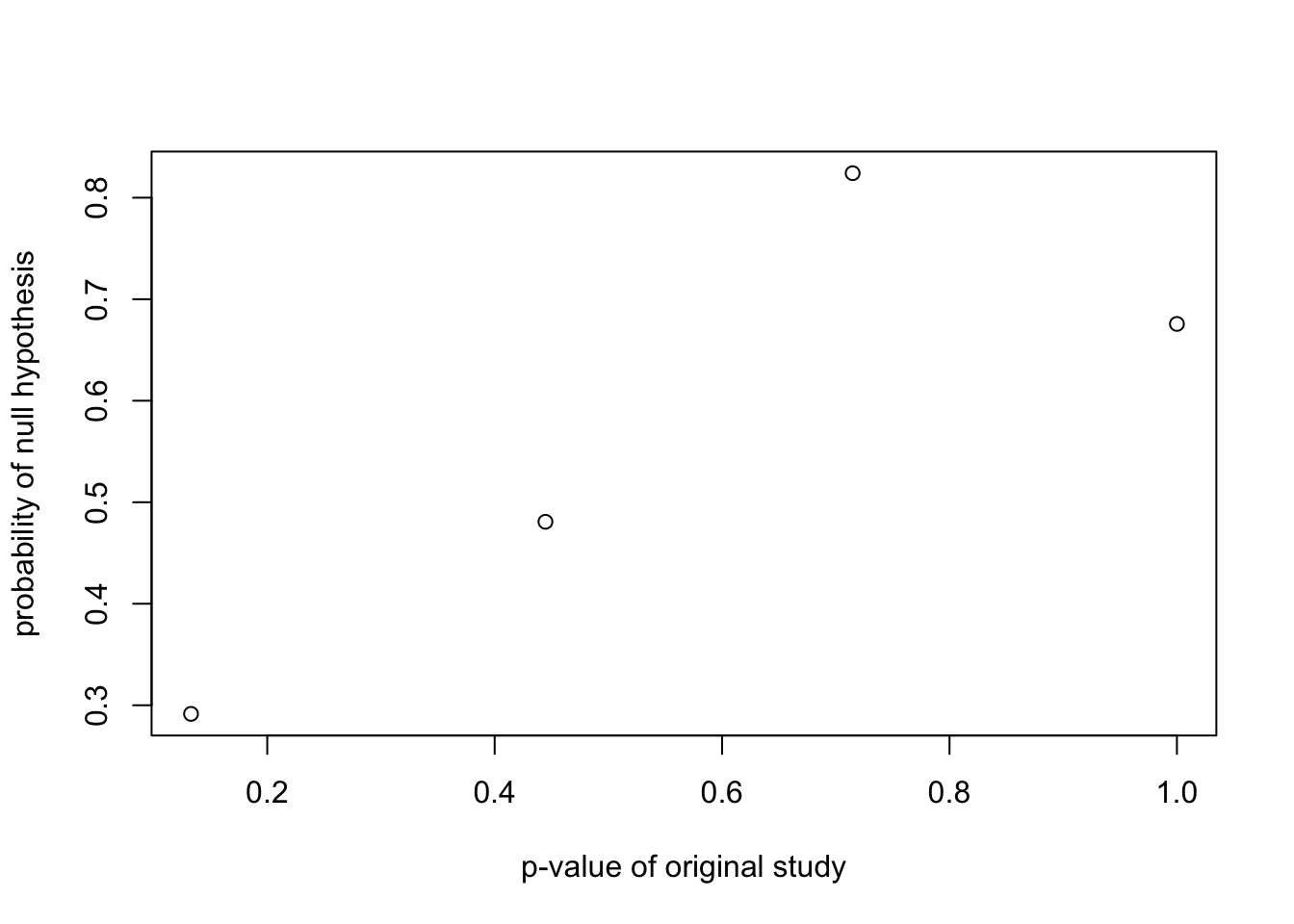
xlab="p-value of original study")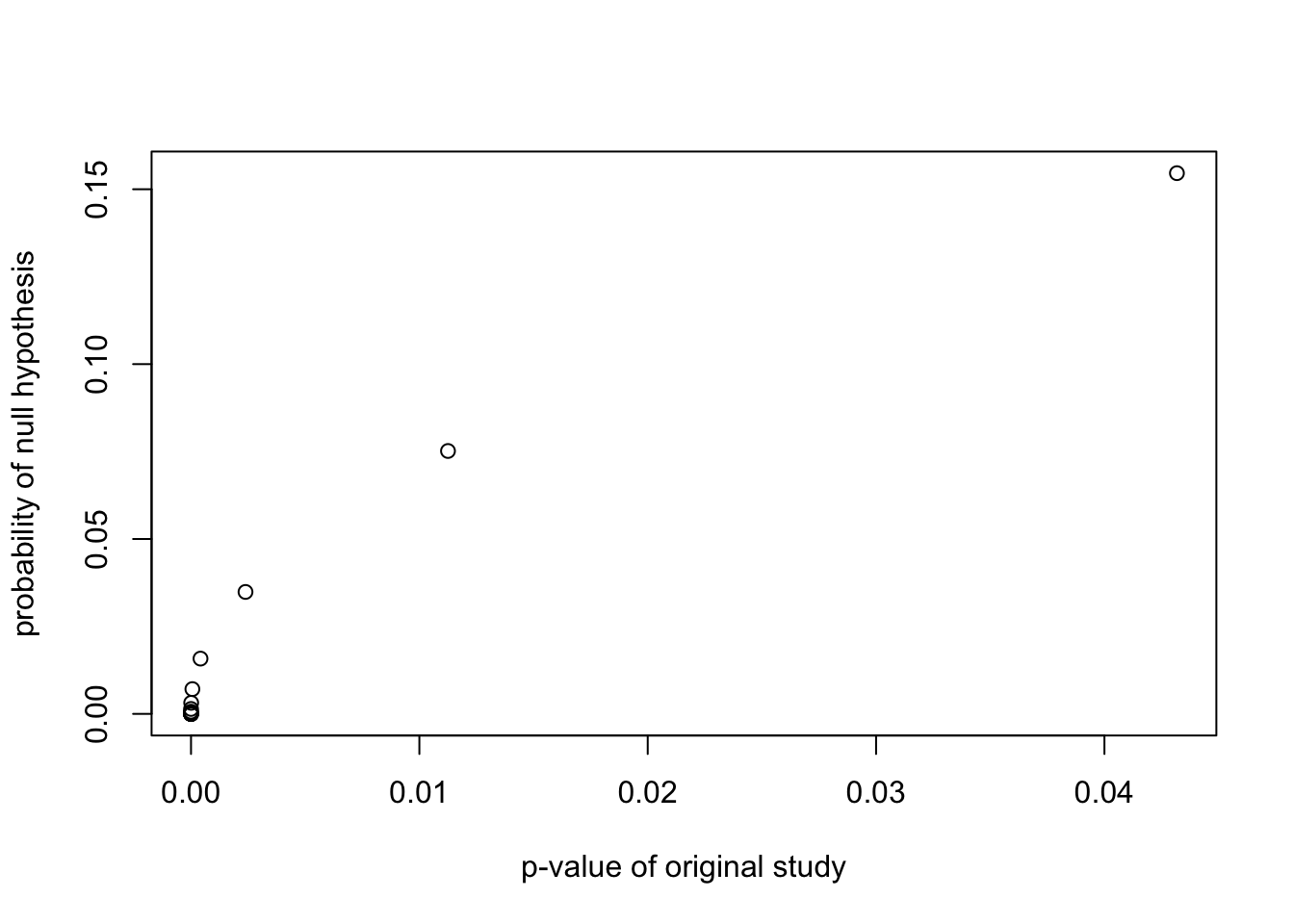
plot(probrep.out$retain$pvalue.retain,
probrep.out$retain$prep.retain,
ylab="probability of replication",
xlab="p-value of original study")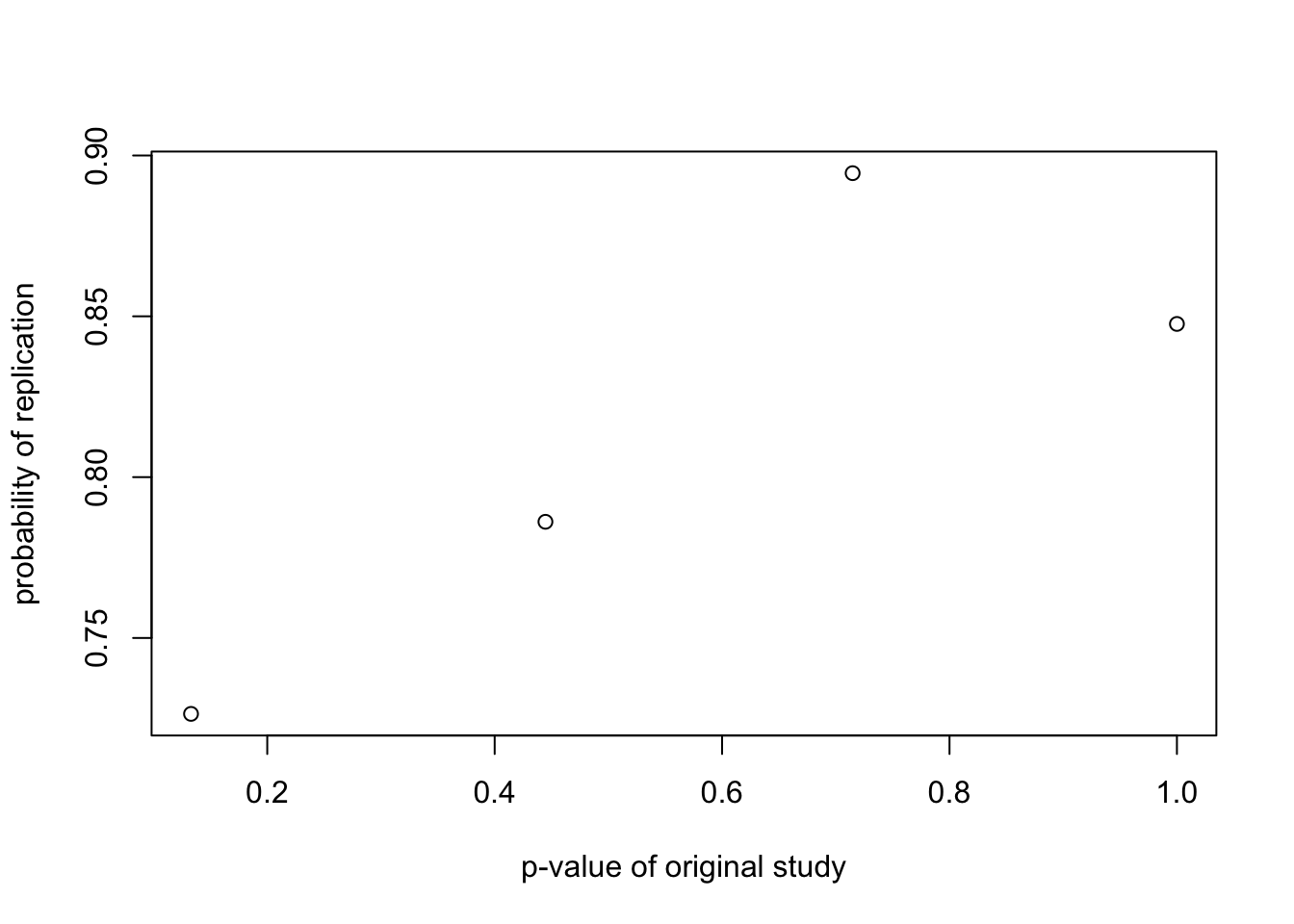
plot(probrep.out$retain$pvalue.retain,
probrep.out$retain$pH0.retain,
ylab="probability of null hypothesis",
xlab="p-value of original study")